Inbreeding and linebreeding
What are inbreeding and linebreeding, and what effect do they have?
In genetic terminology, inbreeding is the breeding of two animals who are related to each other. In its opposite, outcrossing, the two parents are totally unrelated. Since all pure breeds of animal trace back to a relatively limited number of foundation dogs, all pure breeding is by this definition inbreeding, although the term is not generally used to refer to matings where a common ancestor does not occur behind sire and dam in a four or five generation pedigree.
Breeders of purebred livestock have introduced a term, linebreeding, to cover the milder forms of inbreeding. Exactly what the difference is between linebreeding and inbreeding tends to be defined differently for each species and often for each breed within the species. On this definition, inbreeding at its most restrictive applies to what would be considered unquestioned incest in human beings - parent to offspring or a mating between full siblings. Uncle-niece, aunt-nephew, half sibling matings, and first cousin matings are called inbreeding by some people and linebreeding by others.
What does inbreeding (in the genetic sense) do? Basically, it increase the probability that the two copies of any given gene will be identical and derived from the same ancestor. Technically, the animal is homozygous for that gene. The heterozygous animal has some differences in the two copies of the gene Remember that each animal (or plant, for that matter) has two copies of any given gene (two alleles at each locus, if you want to get technical), one derived from the father and one from the mother. If the father and mother are related, there is a chance that the two genes in the offspring are both identical copies contributed by the common ancestor. This is neither good nor bad in itself. Consider, for instance, the gene for PRA (progressive retinal atrophy), which causes progressive blindness. Carriers have normal vision, but if one is mated to another carrier, one in four of the puppies will have PRA and go blind. Inbreeding will increase both the number of affected dogs (bad) and the number of genetically normal dogs (good) at the expense of carriers. Inbreeding can thus bring these undesirable recessive genes to the surface, where they can be removed from the breeding pool.
Unfortunately, we cannot breed animals based on a single gene - the genes come as a package. We may inbreed and rigorously remove pups with PRA or even their parents and littermates from the breeding pool. But remember inbreeding tends to make all genes more homozygous. In at least one breed, an effort to remove the PRA-causing gene resulted in the surfacing of a completely different and previously unsuspected health problem. It is easier and faster to lose genes (sometimes very desirable genes) from the breeding pool when inbreeding is practiced than when a more open breeding system is used. In other words, inbreeding will tend to produce more nearly homozygous animals, but generally some of the homozygous pairs will be "good" and others will be "bad".
Furthermore, there may be genes where heterozygosity is an advantage. There are several variant hemoglobin types in human beings, for instance, where one homozygote suffers from some type of illness, the other homozygote is vulnerable to malaria, and the heterozygote is generally malaria-resistant with little or no negative health impacts from a single copy of the non-standard hemoglobin gene. A more widespread case is the so-called major histocompatibility complex (MHC), a group of genes where heterozygosity seems to improve disease resistance.
Is there a way of measuring inbreeding? Wright developed what is called the inbreeding coefficient. This is related to the probability that both copies of any given gene are derived from the same ancestor. A cold outcross (in dogs, probably a first-generation cross between two purebreds of different, unrelated breeds would be the best approximation) would have an inbreeding coefficient of 0. Note that this dog would not be heterozygous at every locus. There are genes shared with every multicellular organism, genes shared with all animals, genes shared with all animals with backbones, genes shared with all four-limbed animals (including most fish and all amphibians, reptiles, birds and mammals) and with all mammals. Although the DNA might differ slightly, the proteins produced would be functionally the same. Further, the chances are that our dogs with inbreeding coefficient = 0 would still be homozygous for some genes shared by all dogs. The inbreeding coefficient thus specifically refers to those genes that are variable (more than one possible form) in the species and even the breed being considered.
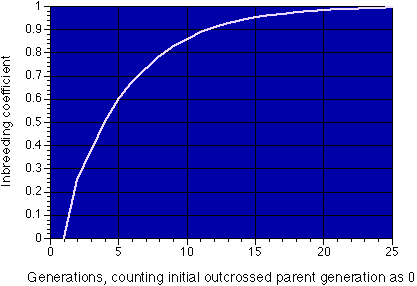
An inbreeding coefficient of 1 (rare in mammals) would result if the only matings practiced over many generations were between full brother and full sister.
The figure shows how the inbreeding coefficient chages with generations of brother-sister matings. As a general rule, this type of mating in domestic animals cannot be kept up beyond 8-10 generations, as by that time the rate of breeding success is very low. However, the rare survivors may go on to found genetically uniform populations.
This has been done in laboratory rodents, producing inbred strains of mice and rats so similar genetically that they easily tolerate skin or organ grafts from other animals from the same inbred strain. However, the process of inbreeding used to create these strains generally results in loss of fertility (first seen in these mammals as a reduction in litter size) which actually kills off the majority of the strains between 8 and 12 generations of this extent of inbreeding. A handful of the initial strains survive this bottleneck, and these are the inbred laboratory strains. However, very little selection other than for viability and fertility is possible during this process. You wind up with animals homozygous for a more or less random selection of whatever genes happened to be in the strains that survived, all of which derive from the parents of the initial pair.
Note that two very inbred parents can produce offspring that have very low inbreeding coefficients if the inbred parents do not have ancestors in common. This, however, assumes that mates are available who are not strongly inbred on a common ancestor. If the parents are related to each other, their own inbreeding coefficients will indeed increase the inbreeding coefficients of their offspring. The critical factor is the coefficient of kinship, which is the inbreeding coefficient of a hypothetical offspring of the two individuals.
Inbreeding has become an important consideration for wildlife conservationists. Many wild populations are in danger of extinction due to some combination of habitat destruction and hunting of the animals, either to protect humans or because the animal parts are considered valuable. (Examples are ivory, rhinorcerus horn, and infant apes for the pet trade, as well as meat hunting.) For some of these animals the only real hope of survival is captive breeding programs. But the number of animals available in such captive breeding programs, especially at a single zoo, is often limited. Biologists are concerned that the resulting inbred populations would not have all of the genes found in the wild populations, and thus lose some flexibility in responding to change. In reaction to this threat they have developed networks such that animals can be exchanged among captive breeding poplulations in such a way as to minimize the overall inbreeding of the captive population. The idea is to select pairs in such a way that the inbreeding coefficient of the offspring is kept as low as possible.
Most elementary genetics books have instructions for calculating the inbreeding coefficient from the pedigree. (For more information, see Dr. Armstrong's site, Significant Relationships.) However, these procedures have two major limitations. First, they are not really designed for cases where there are multiple common ancestors, though they can be used separately for each common ancestor and the results added. Second, they become impossibly complex as the length of the pedigree increases. It is by no means uncommon in dogs, for instance, to have pedigrees which can be researched in the AKC stud book and the KC Gazette and which go back to foundation dogs born around the turn of the century - perhaps 30 or even 40 generations earlier. With this type of long pedigree, foundation animals may appear a million times or more in the pedigree.
With this in mind, a computer program called GENES was developed by Dr. Robert Lacy for the calculation of the inbreeding coefficient, kinship coefficients among animals in the breeding pool, percent contributions of varying founding ancestors, and related output, assuming full pedigrees to the foundation stock were available for all animals currently in the breeding population. For captive breeding populations, the less inbreeding the better, and this is the way the program is used.
In purebred livestock the situtation is a little different - we want homozygosity for those genes which create a desirable similarity to the breed standard. Wright's defense of inbreeding was based on this fact. However, inbreeding tends to remove those heterozygotes which are beneficial (e.g., the MHC) as well as increasing undesirable as well as desirable homozygotes. The practice is most dangerous in the potential increase of homozygous health problems which are not obvious on inspection, but which shorten the life span or decrease the quality of life for the animal.
I do not at the present time have other dog breeds for comparison, but I recently submitted a Shetland Sheepdog pedigree database to Dr. Armstrong for calculation of true inbreeding coefficients. This database was based on full pedigrees of all AKC Shetland Sheepdogs that had sired 10 or more breed champions (males) or produced 5 or more (females.) These top producing animals were set up as the current living population (a somewhat artifial assumption, as the dogs involved where whelped from 1930 to after 1990.) I would love to see some comparisons with other breeds.
Back to Genetics index
sbowling@mosquitonet.comupdated March 8, 2010